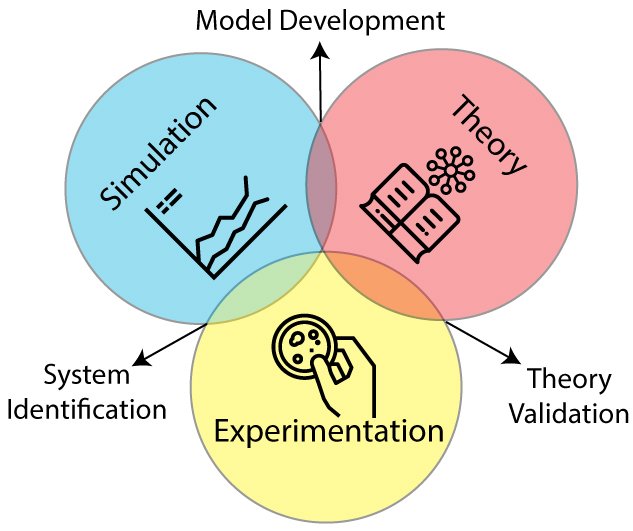
The design foundry of our work is an integration of multidisciplinary methods that propels the development of the lab’s research. It lies in the intersections of theory, simulation, and experimentation. The areas formed by the overlap of these three circles summarize the core methods in the design foundry:
Computation and Theory
Model Development
Use multi-scale models to capture interwoven dynamics across scales in synthetic microbial consortia.
Model bacterial gene expression in stationary phase and complex environments.
Systems modeling beyond synthetic biomolecular networks.
System Identification
Statistical parameter inference methods that utilize multiple data types.
Automated experimental design for system identification by part.
Theory Validation
Dynamical control strategies in natural biological networks.
Implementation and characterization of classic control strategies in synthetic biology.
Publications
Aghlmand, F., Hu, C.Y, Sharma, S., Pochana, K., Murray, R.M, and Emami, A., “A 65nm CMOS Living-Cell Dynamic Fluorescence Sensor with 1.05fA Sensitivity at 600/700nm Wavelengths,” in IEEE Int. Solid-State Circuits Conf., Feb. 2023, Accepted
Hu, C. Y., & Murray, R. M. Layered Feedback Control Overcomes Performance Trade-off in Synthetic Biomolecular Networks. Nat Commun 13, 5393 (2022). https://doi.org/10.1038/s41467-022-33058-6 (Corresponding Author)
Green, L. N., Hu, C. Y., Ren, X. Y., & Murray, R. M. (2019). Bacterial Controller Aided Wound Healing: A Case Study in Dynamical Population Controller Design. bioRxiv,19(7), 3217–15. http://doi.org/10.1101/659714
Hu, C. Y., & Murray, R. M.. Design of a genetic layered feedback controller in synthetic biological circuitry. bioRxiv, 2019. 2(3) 905–911. https://doi.org/10.1101/647057 (Corresponding author)
Hu, C. Y., Takahashi, M. K., Zhang, Y., & Lucks, J. B. (2018). Engineering a Functional Small RNA Negative Autoregulation Network with Model-Guided Design. ACS Synthetic Biology, 7(6), 1507–1518. http://doi.org/10.1021/acssynbio.7b00440
Hu, C. Y., Varner, J. D., & Lucks, J. B. (2015). Generating Effective Models and Parameters for RNA Genetic Circuits. ACS Synthetic Biology, 4(8), 914–926. http://doi.org/10.1021/acssynbio.5b00077